File for givinvg functions of calculating spectrum by Lanczos. More...
#include "Common.h"#include "CalcSpectrumByLanczos.h"#include "Lanczos_EigenValue.h"#include "FileIO.h"#include "wrapperMPI.h"#include "common/setmemory.h"#include "komega/komega.h"#include "mltply.h"#include <mpi.h> Include dependency graph for CalcSpectrumByBiCG.c:
Include dependency graph for CalcSpectrumByBiCG.c:Go to the source code of this file.
Functions | |
| void | ReadTMComponents_BiCG (struct EDMainCalStruct *X, double complex *v2, double complex *v4, double complex *v12, double complex *v14, int Nomega, double complex *dcSpectrum, double complex *dcomega) |
| Read \(\alpha, \beta\), projected residual for restart. More... | |
| int | OutputTMComponents_BiCG (struct EDMainCalStruct *X, int liLanczosStp) |
| write \(\alpha, \beta\), projected residual for restart More... | |
| void | InitShadowRes (struct BindStruct *X, double complex *v4) |
| Initialize Shadow Residual as a random vector (Experimental) More... | |
| int | CalcSpectrumByBiCG (struct EDMainCalStruct *X, double complex *vrhs, double complex *v2, double complex *v4, int Nomega, double complex *dcSpectrum, double complex *dcomega) |
| A main function to calculate spectrum by BiCG method In this function, the \(K\omega\) library is used. The detailed procedure is written in the document of \(K\omega\). https://issp-center-dev.github.io/Komega/library/en/_build/html/komega_workflow_en.html#the-schematic-workflow-of-shifted-bicg-library. More... | |
Detailed Description
File for givinvg functions of calculating spectrum by Lanczos.
Definition in file CalcSpectrumByBiCG.c.
Function Documentation
◆ CalcSpectrumByBiCG()
| int CalcSpectrumByBiCG | ( | struct EDMainCalStruct * | X, |
| double complex * | vrhs, | ||
| double complex * | v2, | ||
| double complex * | v4, | ||
| int | Nomega, | ||
| double complex * | dcSpectrum, | ||
| double complex * | dcomega | ||
| ) |
A main function to calculate spectrum by BiCG method In this function, the \(K\omega\) library is used. The detailed procedure is written in the document of \(K\omega\). https://issp-center-dev.github.io/Komega/library/en/_build/html/komega_workflow_en.html#the-schematic-workflow-of-shifted-bicg-library.
- Return values
-
0 normally finished -1 error
-
Malloc vector for old residual vector ( \({\bf r}_{\rm old}\)) and old shadow residual vector ( \({\bf {\tilde r}}_{\rm old}\)).
-
Set initial result vector(+shadow result vector) Read residual vectors if restart
-
Input \(\alpha, \beta\), projected residual, or start from scratch
-
DO BiCG loop
-
\({\bf v}_{2}={\hat H}{\bf v}_{12}, {\bf v}_{4}={\hat H}{\bf v}_{14}\), where \({\bf v}_{12}, {\bf v}_{14}\) are old (shadow) residual vector.
-
Update projected result vector dcSpectrum.
-
Output residuals at each frequency for some analysis
-
-
END DO BiCG loop
-
Save \(\alpha, \beta\), projected residual
- output vectors for recalculation
- Parameters
-
[in,out] X [in] vrhs [CheckList::idim_max] Right hand side vector, excited state. [in,out] v2 [CheckList::idim_max] Work space for residual vector \({\bf r}\) [in,out] v4 [CheckList::idim_max] Work space for shadow residual vector \({\bf {\tilde r}}\) [in] Nomega Number of Frequencies [out] dcSpectrum [Nomega] Spectrum [in] dcomega [Nomega] Frequency
Definition at line 206 of file CalcSpectrumByBiCG.c.
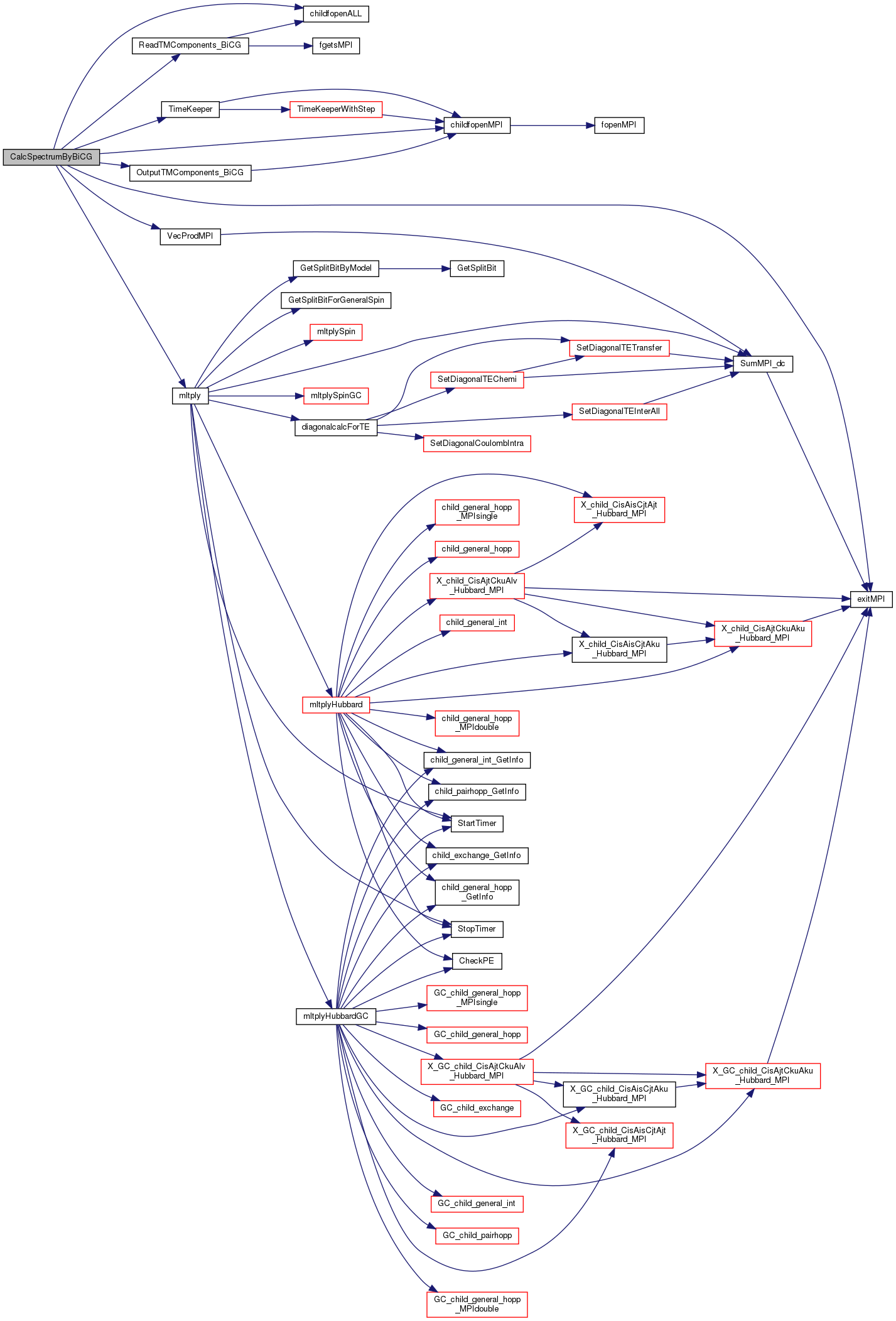
References EDMainCalStruct::Bind, c_GetTridiagonalEnd, c_GetTridiagonalStart, c_InputSpectrumRecalcvecEnd, c_InputSpectrumRecalcvecStart, c_OutputSpectrumRecalcvecEnd, c_OutputSpectrumRecalcvecStart, DefineList::CDataFileHead, cFileNameOutputRestartVec, cFileNameTimeKeep, BindStruct::Check, childfopenALL(), childfopenMPI(), D_FileNameMax, BindStruct::Def, exitMPI(), CheckList::idim_max, DefineList::iFlgCalcSpec, DefineList::Lanczos_max, mltply(), myrank, OutputTMComponents_BiCG(), ReadTMComponents_BiCG(), stdoutMPI, TimeKeeper(), TRUE, and VecProdMPI().
Referenced by CalcSpectrum().
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ InitShadowRes()
| void InitShadowRes | ( | struct BindStruct * | X, |
| double complex * | v4 | ||
| ) |
Initialize Shadow Residual as a random vector (Experimental)
- Parameters
-
[in,out] X [out] v4 [CheckList::idim_max] shadow residual vector
Definition at line 154 of file CalcSpectrumByBiCG.c.
References BindStruct::Check, BindStruct::Def, CheckList::idim_max, DefineList::initial_iv, myrank, nthreads, and VecProdMPI().
 Here is the call graph for this function:
Here is the call graph for this function:◆ OutputTMComponents_BiCG()
| int OutputTMComponents_BiCG | ( | struct EDMainCalStruct * | X, |
| int | liLanczosStp | ||
| ) |
write \(\alpha, \beta\), projected residual for restart
- Parameters
-
[in,out] X [in] liLanczosStp the BiCG step
Definition at line 118 of file CalcSpectrumByBiCG.c.
References EDMainCalStruct::Bind, DefineList::CDataFileHead, cFileNameTridiagonalMatrixComponents, childfopenMPI(), D_FileNameMax, BindStruct::Def, and TRUE.
Referenced by CalcSpectrumByBiCG().
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ ReadTMComponents_BiCG()
| void ReadTMComponents_BiCG | ( | struct EDMainCalStruct * | X, |
| double complex * | v2, | ||
| double complex * | v4, | ||
| double complex * | v12, | ||
| double complex * | v14, | ||
| int | Nomega, | ||
| double complex * | dcSpectrum, | ||
| double complex * | dcomega | ||
| ) |
Read \(\alpha, \beta\), projected residual for restart.
- Parameters
-
[in,out] X [in,out] v2 [CheckList::idim_max] Residual vector [in,out] v4 [CheckList::idim_max] Shadow esidual vector [in,out] v12 [CheckList::idim_max] Old residual vector [in,out] v14 [CheckList::idim_max] Old shadow residual vector [in] Nomega Number of frequencies [in,out] dcSpectrum [Nomega] Projected result vector, spectrum [in] dcomega [Nomega] Frequency
Definition at line 34 of file CalcSpectrumByBiCG.c.
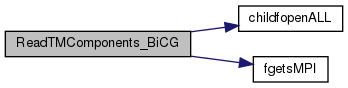
References EDMainCalStruct::Bind, DefineList::CDataFileHead, cFileNameTridiagonalMatrixComponents, BindStruct::Check, childfopenALL(), D_FileNameMax, BindStruct::Def, eps_Lanczos, fgetsMPI(), CheckList::idim_max, DefineList::iFlgCalcSpec, DefineList::Lanczos_max, and stdoutMPI.
Referenced by CalcSpectrumByBiCG().
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function: