Grid search¶
In this section, we will explain how to perform a grid-type search and analyze atomic coordinates from diffraction data.
The grid type search is compatible with MPI. The specific calculation procedure is the same as for minsearch.
However, it is necessary to prepare the data MeshData.txt to give the search grid in advance.
Location of the sample files¶
The sample files are located in sample/sim-trhepd-rheed/single_beam/mapper.
The following files are stored in the folder
bulk.txtInput file of
bulk.exeexperiment.txt,template.txtReference file to proceed with calculations in the main program.
ref_ColorMap.txtA file to check if the calculation was performed correctly (the answer to
ColorMap.txtobtained by doing this tutorial).input.tomlInput file of the main program.
prepare.sh,do.shScript prepared for bulk calculation of this tutorial.
Below, we will describe these files and then show the actual calculation results.
Reference file¶
The template.txt and experiment.txt are the same as in the previous tutorial (Nealder-Mead optimization).
However, to reduce the computation time, the value is fixed to 3.5 instead of value_03, and the grid is searched in 2D.
The actual grid to be searched is given in MeshData.txt.
In the sample, the contents of MeshData.txt are as follows.
1 6.000000 6.000000
2 6.000000 5.750000
3 6.000000 5.500000
4 6.000000 5.250000
5 6.000000 5.000000
6 6.000000 4.750000
7 6.000000 4.500000
8 6.000000 4.250000
9 6.000000 4.000000
...
The first column is the serial number, and the second and subsequent columns are the values of value_0 , value_1 that go into template.txt, in that order.
Input file¶
This section describes the input file for the main program, input.toml.
The details of input.toml can be found in the input file.
The following is the content of input.toml in the sample file.
[base]
dimension = 2
[solver]
name = "sim-trhepd-rheed"
[solver.config]
calculated_first_line = 5
calculated_last_line = 74
row_number = 2
[solver.param]
string_list = ["value_01", "value_02" ]
degree_max = 7.0
[solver.reference]
path = "experiment.txt"
first = 1
last = 70
[algorithm]
name = "mapper"
label_list = ["z1", "z2"]
First, [base] section is explained.
The
dimensionis the number of variables to be optimized, in this case2since we are optimizing two variables as described intemplate.txt.
The [solver] section specifies the solver to be used inside the main program and its settings.
The
nameis the name of the solver you want to use, which in this tutorial issim-trhepd-rheed, since we will be using it for our analysis.
The solver can be configured in the subsections [solver.config], [solver.param], and [solver.reference].
The [solver.config] section specifies options for reading the output file produced by the main program’s internal call, surf.exe.
The
calculated_first_linespecifies the first line to read from the output file.The
calculated_last_linespecifies the last line of the output file to be read.The
row_numberspecifies the number of columns in the output file to read.
The [solver.param] section specifies options for reading the output file produced by the main program’s internal call, surf.exe.
The
string_listis a list of variable names to be read intemplate.txt.degree_maxspecifies the maximum angle in degrees.
The [solver.reference] section specifies the location of the experimental data and the range to read.
The
pathspecifies the path where the experimental data is located.The
firstspecifies the first line of the experimental data file to read.The
endspecifies the last line of the experimental data file to read.
The [algorithm] section specifies the algorithm to use and its settings.
The
nameis the name of the algorithm you want to use, in this tutorial we will usemappersince we will be using grid-search method.The
label_listis a list of label names to be attached to the outputvalue_0x(x=1,2).
For details on other parameters that can be specified in the input file, please see the Input File chapter.
Calculation execution¶
First, move to the folder where the sample files are located (we will assume that you are directly under the directory where you downloaded this software).
cd sample/sim-trhepd-rheed/single_beam/minsearch
Copy bulk.exe and surf.exe.
cp ../../../../../sim-trhepd-rheed/src/TRHEPD/bulk.exe .
cp ../../../../../sim-trhepd-rheed/src/TRHEPD/surf.exe .
First, run bulk.exe to create bulkP.b.
./bulk.exe
After that, run the main program (the computation time takes only a few seconds on a normal PC).
mpiexec -np 2 python3 ../../../../src/py2dmat_main.py input.toml | tee log.txt
Here, the calculation using MPI parallel with 2 processes will be done.
When executed, a folder for each rank will be created, and a subfolder Log%%%%% (where %%%%% is the grid id) will be created under it.
(The grid id is assigned to the number in MeshData.txt).
The standard output will be seen like this.
Iteration : 1/33
Read experiment.txt
mesh before: [1.0, 6.0, 6.0]
z1 = 6.00000
z2 = 6.00000
[' 6.00000', ' 6.00000']
PASS : degree in lastline = 7.0
PASS : len(calculated_list) 70 == len(convolution_I_calculated_list)70
R-factor = 0.04785241875354398
...
The z1 and z2 are the candidate parameters for each mesh and the R-factor at that time.
Finally, the R-factor calculated for all the points on the grid will be output to ColorMap.txt.
In this case, the following results will be obtained.
6.000000 6.000000 0.047852
6.000000 5.750000 0.055011
6.000000 5.500000 0.053190
6.000000 5.250000 0.038905
6.000000 5.000000 0.047674
6.000000 4.750000 0.065919
6.000000 4.500000 0.053675
6.000000 4.250000 0.061261
6.000000 4.000000 0.069351
6.000000 3.750000 0.071868
6.000000 3.500000 0.072739
...
The first and second columns will contain the values of value_01 and value_02, and the third column will contain the R-factor.
Note that do.sh is available as a script for batch calculation.
In do.sh, it also compares the difference between res.txt and ref.txt.
Here is what it does, without further explanation.
sh prepare.sh
./bulk.exe
time mpiexec -np 2 python3 ../../../../src/py2dmat_main.py input.toml
echo diff ColorMap.txt ref_ColorMap.txt
res=0
diff ColorMap.txt ref_ColorMap.txt || res=$?
if [ $res -eq 0 ]; then
echo TEST PASS
true
else
echo TEST FAILED: ColorMap.txt and ref_ColorMap.txt differ
false
fi
Visualization of calculation results¶
By seeing ColorMap.txt, we can estimate the region where the small parameters of R-factor are located.
In this case, the following command will create a two-dimensional parameter space diagram ColorMapFig.png.
python3 plot_colormap_2d.py
Looking at the generated figure, we can see that it has a minimum value around (5.25, 4.25).
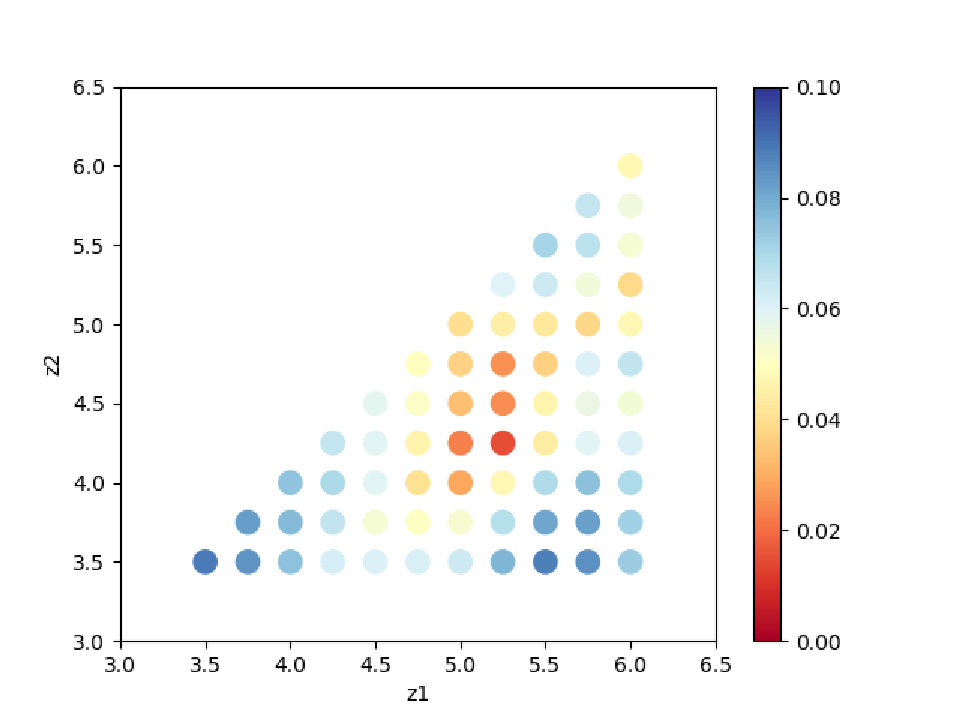
Fig. 3 R-factor on a two-dimensional parameter space.¶
RockingCurve.txt is stored in each subfolder.
By using it, you can compare the results with the experimental values following the procedure in the previous tutorial.