3.4. Number density of the hardcore Bosons on a square lattice¶
In this tutorial, we will calculate the chemical potential dependence of the number density of the hardcore Bose-Hubbard model with the nearest neighbor repulsive on a \(8\times8\) square lattice.
The following Python script (sample/dla/03_bosesquare/exec.py) performs DSQSS/DLA work-flow for each parameter (chemical potential) automatically.
import subprocess
from dsqss.dla_pre import dla_pre
from dsqss.result import Results
V = 3
L = [8, 8]
beta = 10.0
lattice = {"lattice": "hypercubic", "dim": 2, "L": L}
hamiltonian = {"model": "boson", "t": 1, "V": V, "M": 1}
parameter = {"beta": beta, "nset": 4, "ntherm": 100, "ndecor": 100,
"nmcs": 100, "wvfile": "wv.xml", "dispfile": "disp.xml"}
name = "amzu"
mus = [-4.0, -2.0, 0.0, 2.0, 2.5, 3.0, 6.0, 9.0, 9.5, 10.0, 12.0, 14.0]
output = open("{}.dat".format(name), "w")
for i, mu in enumerate(mus):
ofile = "res_{}.dat".format(i)
pfile = "param_{}.in".format(i)
sfoutfile = "sf_{}.dat".format(i)
cfoutfile = "cf_{}.dat".format(i)
ckoutfile = "ck_{}.dat".format(i)
hamiltonian["mu"] = mu
parameter["outfile"] = ofile
parameter["sfoutfile"] = sfoutfile
parameter["cfoutfile"] = cfoutfile
parameter["ckoutfile"] = ckoutfile
dla_pre(
{"parameter": parameter, "hamiltonian": hamiltonian, "lattice": lattice}, pfile
)
cmd = ["dla", pfile]
subprocess.call(cmd)
res = Results(ofile)
output.write("{} {}\n".format(mu, res.to_str(name)))
output.close()
Before executing this script, source a configuring file dsqssvars-VERSION.sh in order to set environment variables
(replace VERSION with the version of DSQSS, e.g., 2.0.0.)
$ source $DSQSS_INSTALL_DIR/share/dsqss/dsqssvars-VERSION.sh
$ python exec.py
The result is written to amzu.dat (Fig. 3.2).
You can see a density plateau around \(\mu=6\) . In this region, a checker board solid phase due to repulsive interaction appears.
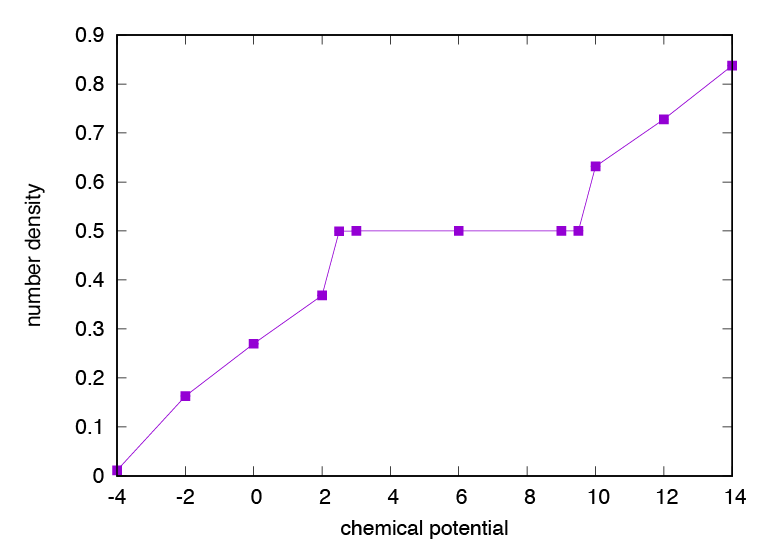
Fig. 3.2 Chemical potential dependence of number density of repulsive hardcore bosons.¶
